Clinical utility of endoscopic ultrasound-guided tissue acquisition for comprehensive genomic profiling of pancreatic cancer
Article information
Abstract
Background/Aims
Endoscopic ultrasound-guided tissue acquisition (EUS-TA) is essential for the diagnosis of pancreatic cancer. The feasibility of comprehensive genomic profiling (CGP) using samples obtained by EUS-TA has been under recent discussion. This study aimed to evaluate the utility of EUS-TA for CGP in a clinical setting.
Methods
CGP was attempted in 178 samples obtained from 151 consecutive patients with pancreatic cancer at the Aichi Cancer Center between October 2019 and September 2021. We evaluated the adequacy of the samples for CGP and determined the factors associated with the adequacy of the samples obtained by EUS-TA retrospectively.
Results
The overall adequacy for CGP was 65.2% (116/178), which was significantly different among the four sampling methods (EUS-TA vs. surgical specimen vs. percutaneous biopsy vs. duodenal biopsy, 56.0% [61/109] vs. 80.4% [41/51] vs. 76.5% [13/17] vs. 100.0% [1/1], respectively; p=0.022). In a univariate analysis, needle gauge/type was associated with adequacy (22 G fine-needle aspiration vs. 22 G fine-needle biopsy [FNB] vs. 19 G-FNB, 33.3% (5/15) vs. 53.5% (23/43) vs. 72.5% (29/40); p=0.022). The sample adequacy of 19 G-FNB for CGP was 72.5% (29/40), and there was no significant difference between 19 G-FNB and surgical specimens (p=0.375).
Conclusions
To obtain adequate samples for CGP with EUS-TA, 19 G-FNB was shown to be the best in clinical practice. However, 19 G-FNB was not still sufficient, so further efforts are required to improve adequacy for CGP.
INTRODUCTION
Pancreatic cancer is one of the deadliest cancers worldwide. According to the latest cancer statistics from the Cancer Information Service of the National Cancer Center,1 pancreatic cancer is the fourth leading cause of cancer-related deaths, with a mortality rate of 30.1 per 100,000 men and 28.7 per 100,000 women in Japan in 2019. A study conducted in the United States in 2019 estimated that there would be 56,770 new cases that year, and that 45,750 patients would die from the disease.2 The reason for the poor prognosis is that surgical resection is the only curative treatment and it is difficult to diagnose pancreatic cancer at an early stage. In addition, there are limited effective therapies available.
However, prognosis is expected to improve following the recent introduction of “precision medicine”, which is based on tumor genomic profiling. The recent National Comprehensive Cancer Network guidelines recommend that germline testing, gene profiling, and mismatch repair/microsatellite instability testing should be considered for metastatic pancreatic ductal cancer. Japanese health insurance has covered comprehensive genomic profiling (CGP) since June 2019. The Japanese Ministry of Health, Labour and Welfare has approved two CGP tests for the use in cancer diagnosis and it have been implemented in routine clinical practice in Japan; the FoundationOne CDx (CDx; Foundation Medicine Inc., Cambridge, MA, USA) and OncoGuide NCC Oncopanel (NCC) tests (Sysmex, Kobe, Japan).3 The Japanese government appointed our center as a hub for cancer genome medicine in September 2019, and we have been performing CGP at this capacity since October 2019.4
Endoscopic ultrasound-guided tissue acquisition (EUS-TA) via fine-needle aspiration (FNA) or fine-needle biopsy (FNB) is a useful and indispensable technique in the diagnosis of pancreatic cancer.5 Recent reports have demonstrated the usefulness of genomic testing of samples obtained using EUS-TA. Elhanafi et al.6 used a 47-gene comprehensive solid tumor panel and Takano et al.7 performed next-generation sequencing of 50 cancer-related genes. In both cases, the samples obtained using EUS-TA were sufficient for the custom panel analysis. Although results have been reported in experimental studies, few studies have investigated the usefulness and potential of EUS-TA for obtaining samples for CGP utilizing FoundationOne CDx and OncoGuide NCC Oncopanel tests. Kandel et al.8 report that EUS-FNB was superior to EUS-FNA, in providing adequate samples for CDx testing. However, it is unknown whether the results of these feasibility studies are applicable in clinical settings. This study aimed to evaluate the utility of EUS-TA in clinical practice to obtain samples for CGP and to clarify the optimal method of EUS-TA for adequate CGP samples.
METHODS
Patients
We attempted CGP for 178 samples obtained from 151 consecutive patients with pancreatic cancer at the Aichi Cancer Center, Japan, between October 2019 and September 2021. Clinical data were collected retrospectively for these 151 patients.
EUS procedure
During the procedure the patient was in the left lateral position under conscious sedation with intravenous administration of midazolam and pethidine. EUS-TA was performed using a convex echo-endoscope (GU-UCT260; Olympus Medical Systems, Tokyo, Japan or EG-580UT; Fujifilm, Tokyo, Japan). The type and size of the needle were selected at the discretion of the endosonographer among 19-, 22-, or 25-gauge needles (FNA needle; EZ shot 3 plus; Olympus Medical Systems or FNB needle; Acquire; Boston Scientific, MA, USA). Color Doppler imaging was performed during and after EUS-TA to avoid blood vessels and identify signs of bleeding (newly hypoechoic or hyperechoic areas). Negative pressure was used with a 20 mL syringe in some cases, but never in patients with an increased risk of bleeding, for example, in cases where a 19 G-FNB needle was used, or in hypervascular tumors. It has been reported that the non-suction technique is preferred to avoid blood contamination.9 In cases of unresectable pancreatic cancer, we performed EUS-TA preferentially using a 19 G-FNB considering CGP at the time of diagnosis. We performed re-EUS-TA when CGP was unsuccessful, although a diagnosis was made. We also used 19 G-FNB in these cases (Fig. 1). We used rapid on-site evaluation (ROSE) to confirm that the sample tissue had been obtained. If ROSE showed an insufficient biopsy sample, one or two additional punctures were performed, and the samples were placed directly in formalin. In cases where ROSE were not possible, we performed three punctures and placed the samples directly in formalin.

Adequate and inadequate histologic specimens obtained by EUS-FNB in the same patient (hematoxylin and eosin stain). (A) Initial inadequate sample obtained with one puncture, using a 22 G-FNB needle with suction, and processed in cell block. Although this sample was sufficient for diagnosis, it did not meet the prescreening criteria for comprehensive genomic profiling due to insufficient material. (B) Adequate sample obtained at re-EUS-TA with four punctures, using a 19 G-FNB needle without suction, placed in formalin directly and processed in biopsy. The sample could be analyzed by FoundationOne CDx (Foundation Medicine Inc.). EUS, endoscopic ultrasound; FNB, fine-needle biopsy; TA, tissue acquisition.
Adverse events were graded according to the lexicon of the American Society for Gastrointestinal Endoscopy.10 Significant bleeding events were defined as follows: >2 g/dL decrease in hemoglobin levels compared to baseline and/or a history of melena, hematemesis, or hematochezia with no other cause of upper gastrointestinal bleeding. Nonsignificant bleeding events were defined as follows: hematoma and/or bleeding that could be confirmed on EUS or endoscopic imaging during EUS-TA without a >2 g/dL decrease in hemoglobin levels.
Percutaneous biopsy
Radiologists performed percutaneous biopsy using an 18 G-true cut needle. They collected four to five samples, which were placed directly in formalin without ROSE.
Histologic evaluation
In the prescreening for CGP, a pathologist at our institution evaluated the adequacy of the samples in terms of the overall amount of tissue and the proportion of tumor nuclei.
According to the CDx test guidelines,11 formalin-fixed paraffin-embedded (FFPE) specimens are used in molecular analysis. Sample adequacy was defined according to the following criteria: (1) a sample size of either one block (FFPE) and one hematoxylin and eosin (H&E)-stained slide, or ten unstained slides (positively charged and unbaked at 4 to 5 µm thick) and one hematoxylin and eosin-stained slide; (2) FFPE specimen surface area of ≥25 mm2 (5×5 mm); and (3) percentage of tumor nuclei ≥20% (defined as the total number of tumor cells divided by the total number of all cells with nuclei).
For the NCC Oncopanel test sample adequacy was defined as (1) five unstained slides (10 µm thick), FFPE specimen surface area of 16 mm2 (4×4 mm), and (2) percentage tumor nuclei ≥20%.12 In the prescreening, samples with low tumor cellularity (defined as percentage of tumor nuclei ≤20%) or insufficient material, were judged inadequate. Submission of samples for CGP were based on the suggestions from the pathologist. The type of CGP was also determined by the pathologist. If CDx was possible, we prioritized CDx over NCC Oncopanel, because CDx is also a companion diagnostic test.
Inadequacy of the pathological sample for CGP was defined as prescreening for CGP judged inadequate or sample unanalyzable by CGP. Adequacy of the sample was defined as analyzable CGP (CDx or NCC Oncopanel).
Statistical analysis
Categorical parameters were compared using the chi-squared or Fisher’s exact tests. Two-sided p-values of <0.05 were considered significant. All statistical analyses were performed using the StatMate V statistical software (ATMS, Tokyo, Japan).
Ethical statements
All patients provided informed consent for the procedure, and institutional review board of Aichi Cancer Center Hospital approved this study (approval number: 2021-0-154).
RESULTS
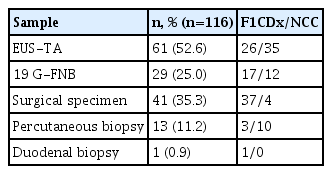
During the study period, CGP was attempted in 151 patients. Patient characteristics are shown in Table 1. The median age was 65 years (range, 23–79 years), and 51.0% of patients were men (77 men and 74 women). The most common histological type was pancreatic ductal adenocarcinoma (85.4%). CGP was attempted in 178 samples from the patient group. The sample adequacy for CGP for each sampling method is shown in Table 2. Most samples were obtained using EUS-TA (61.2%), followed by surgical specimens, percutaneous biopsy and duodenal biopsy. Overall, 65.2% (116/178) of the samples were adequate for CGP. There was a significant difference in the adequacy the of samples among the four groups (EUS-TA vs. surgical specimen vs. percutaneous biopsy vs. duodenal biopsy: 56.0% (61/109) vs. 80.4% (41/51) vs. 76.5% (13/17) vs. 100.0% (1/1), respectively; p=0.022) The reasons for the sample inadequacies are shown in Table 3. In EUS-TA, 72.9% of the inadequate samples had insufficient material, whereas in the surgical specimens, 60.0% of the inadequate samples had low tumor cellularity. In one surgical specimen with insufficient material, the patient underwent a trial laparotomy with dissemination resection.
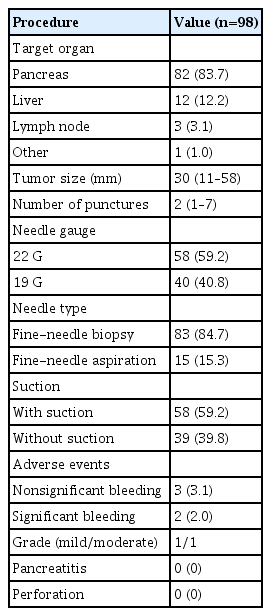
Of the 109 samples obtained by EUS-TA and CGP attempted, we excluded 13 cases for the use of an irregular needle type, which was only used in a few cases, and cases that were performed at another hospital. Finally, we included 98 samples obtained by EUS-TA. The details of the EUS-TA procedures are presented in Table 4. The pancreas was the most common target organ, and FNB was used in 84.7% of the EUS-TA cases. Adverse events occurred in five patients, all of whom had bleeding following FNB. There were three cases of nonsignificant bleeding that did not require any intervention and two cases of significant bleeding. In one of these cases, the bleeding was mild, with a >2 g/dL decrease in the hemoglobin level compared with baseline, and the patient was monitored for one extra night as an inpatient. In the other case, EUS-TA was performed with 19 G-FNB with two punctures, and the bleeding was moderate (melena and hematemesis). Endoscopic examination and contrast-enhanced computed tomography imaging revealed no evidence of active bleeding, and a blood transfusion was performed after which the patient improved.
The results of the factors associated with adequacy for CGP (CDx and NCC) in the 98 samples are shown in Table 5. Median tumor size was analyzed separately. In the univariate analysis, the needle gauge/type was associated with adequacy (22 G-FNA vs. 22 G-FNB vs. 19 G-FNB; 33.3% (5/15) vs. 53.5% (23/43) vs. 72.5% (29/40); p=0.022). There was no significant difference between 19 G-FNB and surgical specimen in terms of sample adequacy (72.5% vs. 80.3%, p=0.375).
CGP and mutation profile
Of the total patient group, 116 (76.8%) underwent CGP in a clinical setting. The details of the CGP type are listed in Table 6. EUS-TA was used to obtain 52.6% of CGP samples in patients with pancreatic cancer. In these EUS-TA samples, CDx was performed in 42.6% (26/61). The CDx test was performed in 58.6% (17/29) of the 19 G-FNB cases.
Figure 2 shows the frequency of major driver genes among the 97 cases of pancreatic ductal adenocarcinoma: KRAS mutations were detected in 92.7%, TP53 mutations in 72.2%, SMAD4 mutations in 34.0%, and CDKN2A mutations in 29.9%. Mutations in any of these four major driver genes were found in 94.8% of cases of pancreatic ductal adenocarcinoma.
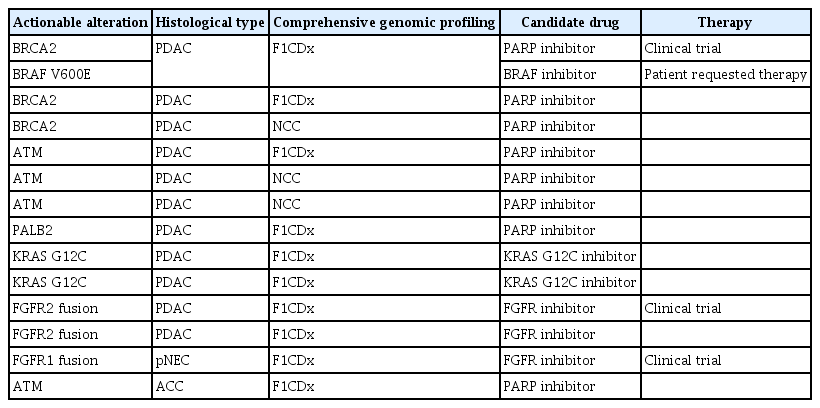
The median overall number of mutations was three (range, 0–20). Thirteen patients (11.2%, 13/116) had actionable alterations, one patient had high microsatellite instability, and four patients (3.4%, 4/116) received treatment based on CGP. The results of actionable alterations are listed in Tables 7 and 8.
DISCUSSION
Pancreatic ductal adenocarcinoma is one of the most lethal solid malignancies. Precision medicine is expected to improve treatment outcomes and prognoses. Research has identified KRAS, TP53, CDKN2A, and SMAD4 as major driver genes for pancreatic cancer,13,14 but there has been low detection of other gene alterations.15 Similarly, in the present study, mutations in these four major driver genes were found in 94.8% of pancreatic ductal adenocarcinomas. There has been little recent progress in the development of drugs for these major driver genes, but improvements are expected with the development of precision medicine and tailored therapeutic strategies.
Our study demonstrated the adequacy of samples obtained using EUS-TA for CGP in a clinical setting. Samples from 116 patients (76.8%) underwent CGP, and approximately 50% of CGP for pancreatic cancer used samples obtained by EUS-TA. EUS-TA plays an important role in the diagnosis of pancreatic cancer and also an increasingly important role in precision medicine. Therefore, it is of great clinical importance that the samples obtained are adequate for CGP.
Song et al.16 report the results of a randomized controlled trial that compared EUS-FNA (without ROSE) using 19 G vs. 22 G needles in 117 patients. The diagnostic accuracy was similar between the needles. However, the 19 G needle provided more cellular material with fewer passes than the 22 G needle (2.4 vs. 2.8; respectively; p=0.01) and no complications were observed in either of the groups. The European Society of Gastrointestinal Endoscopy guidelines17 also state that for EUS-FNA of pancreatic lesions, 19, 22, and 25 G needles are characterized by similar diagnostic yields and safety profiles. However, in the present study, 19 G needles provided more cellular material than thinner needles. A recent meta-analysis of EUS-FNA versus EUS-FNB18 reported no significant difference in diagnostic yield between FNA and FNB when FNA was accompanied by ROSE. However, in the absence of ROSE, FNB is associated with relatively better diagnostic adequacy for solid pancreatic lesions. FNB also requires fewer passes to establish the diagnosis. Studies on the diagnosis of autoimmune pancreatitis19 and subepithelial lesions20 report that EUS-FNB is superior to EUS-FNA. Several other studies have also reported on the efficiency of EUS-FNB for histological specimen acquisition and sampling.21-23
For the sole purpose of diagnosing pancreatic cancer, we believe that any gauge and type of needle can be used. However, CGP requires numerous samples and is a fairly expensive examination. Considering the patient’s disadvantage if re-EUS-TA is required and the time consumed in the event of failure, failure should be avoided. Based on available research on the various methods for EUS-TA, we believe that 19 G-FNB provides the best sample size. Therefore, we have introduced the use of 19 G-EUS-FNB for unresectable cases. In our study, 19 G-FNB had the greatest effect on the adequacy of the obtained specimens. In addition, no severe adverse events were associated with EUS-TA. Even when what we believe to be the best method (19 G-FNB) was used during EUS-TA for CGP, the adequacy rate was 72.5%. No significant difference was found when implementing 19 G-FNB in surgical specimen; however, further effort is needed to increase the adequacy rate of EUS-TA.
Park et al.24 report that the use of a large-gauge needle is an independent factor associated with successful CGP. Kandel et al.8 report that the adequacy of specimens obtained by 19-22 G EUS-FNB for CDx was 78%, but this was a feasibility study. In the present study, the adequacy of specimens obtained by EUS-TA was 56.0% (61/109), and the adequacy reached 72.5% (29/40) for 19 G-FNB. However, only 50% of the adequate samples obtained by 19 G-FNB could be analyzed using CDx. We believe that this discrepancy occurred because of the strict prescreening criteria since we wanted to avoid submitting unanalyzable samples for CGP, which is a treatment that is covered by medical insurance. Of the 62 inadequate samples, 93.5% (58/62) were inadequate at prescreening, and only four samples were unanalyzable for CGP.
Larson et al.25 report that metastatic samples were more likely to be adequate for next-generation sequencing of pancreatic exocrine malignancies than pancreatic samples (p=0.0357). Metastatic samples were included in the percutaneous biopsy samples in the present study, which suggests that metastatic tumors amenable to percutaneous biopsy using large needles can provide a high yield of tumor tissue adequate for next-generation sequencing. Our results relate to EUS-TA only and showed no significant difference in target; however, metastatic samples had high adequacy. It may be advisable to consider EUS-TA for metastatic as well as primary lesions for CGP.
Our study had several limitations. First, it was a retrospective study performed at a single center. Second, because the samples for CGP was surplus after they were used for diagnosis, the samples in our study were potentially affected by a selection bias. Nonetheless, our study demonstrates the importance of EUS-TA using 19 G-FNB for successful CGP in a clinical setting. Third, because we used two different genomic profiling tests, the conditions were not identical. However, this study was conducted under the same conditions as the EUS-TA for CGP in clinical practice.
In conclusion, this is the first study to evaluate the utility of EUS-TA for CGP in clinical settings. Our results suggest that, among the EUS-TA techniques used to obtain samples for CGP, the most promising results were obtained using 19 G-FNB. Therefore, the 19 G-FNB is the first choice when considering CGP. However, even with the use of 19 G-FNB, the technique is not perfect. Considering the patient’s disadvantage if re-EUS-TA is required and the time consumed in the event of failure, further efforts are needed to increase the adequacy rate in EUS-TA. The development of precision medicine will improve treatment outcomes and prognosis in pancreatic cancer; therefore, it is important that adequate specimens are obtained to undergo CGP for unresectable pancreatic cancer.
Notes
Conflicts of Interest
Dr. Mizuno reports the following grants, none of which are connected to the submitted work: from Taiho Pharmaceutical, during the conduct of the study; grants and personal fees from Novartis; personal fees from AstraZeneca; grants from NanoCarrier; grants and personal fees from MSD; grants from Dainippon Sumitomo Pharma; grants from ASLAN Pharmaceuticals; grants from Incyte; grants from Ono Pharmaceutical; personal fees from Teijin Pharma; grants and personal fees from Yakult Honsha; personal fees from Fujifilm Toyama Chemical; and grants from Seagen.
Funding
None.
Author Contributions
Conceptualization: NO, KH; Data curation: NO; Investigation: NO; Methodology: KH; Project administration: KH; Resources: NO, SH, TK, YK, DF, TY; Supervision: KH; Visualization: NO; Writing–original draft: NO; Writing–review & editing: all authors.