Performance comparison between two computer-aided detection colonoscopy models by trainees using different false positive thresholds: a cross-sectional study in Thailand
Article information
Abstract
Background/Aims
This study aims to compare polyp detection performance of “Deep-GI,” a newly developed artificial intelligence (AI) model, to a previously validated AI model computer-aided polyp detection (CADe) using various false positive (FP) thresholds and determining the best threshold for each model.
Methods
Colonoscopy videos were collected prospectively and reviewed by three expert endoscopists (gold standard), trainees, CADe (CAD EYE; Fujifilm Corp.), and Deep-GI. Polyp detection sensitivity (PDS), polyp miss rates (PMR), and false-positive alarm rates (FPR) were compared among the three groups using different FP thresholds for the duration of bounding boxes appearing on the screen.
Results
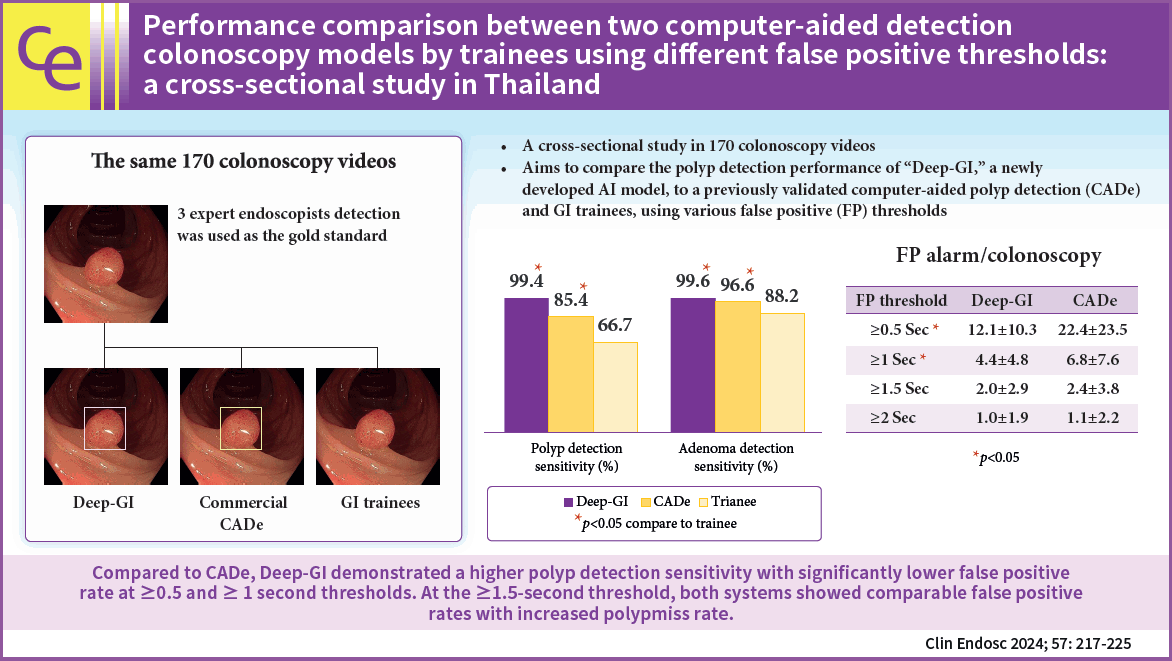
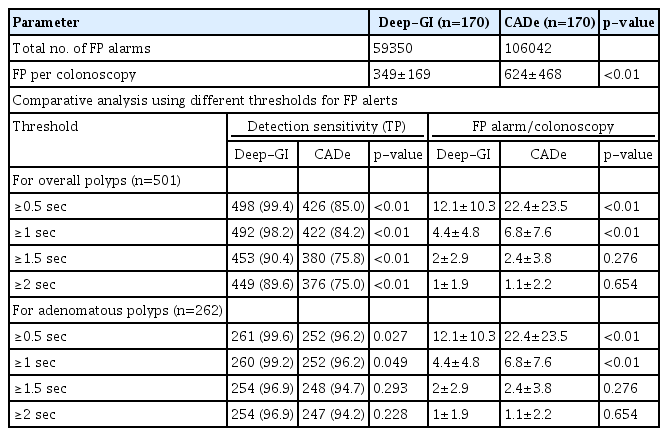
In total, 170 colonoscopy videos were used in this study. Deep-GI showed the highest PDS (99.4% vs. 85.4% vs. 66.7%, p<0.01) and the lowest PMR (0.6% vs. 14.6% vs. 33.3%, p<0.01) when compared to CADe and trainees, respectively. Compared to CADe, Deep-GI demonstrated lower FPR at FP thresholds of ≥0.5 (12.1 vs. 22.4) and ≥1 second (4.4 vs. 6.8) (both p<0.05). However, when the threshold was raised to ≥1.5 seconds, the FPR became comparable (2 vs. 2.4, p=0.3), while the PMR increased from 2% to 10%.
Conclusions
Compared to CADe, Deep-GI demonstrated a higher PDS with significantly lower FPR at ≥0.5- and ≥1-second thresholds. At the ≥1.5-second threshold, both systems showed comparable FPR with increased PMR.
INTRODUCTION
Colorectal cancer (CRC) is the world's third leading cause of cancer-related death.1,2 Over the past decade, CRC incidence and mortality have declined due to an increase in CRC screening and other preventive examinations.3 Among screening tools, colonoscopy has become the gold standard because of its ability to detect and remove premalignant colorectal polyps. It is estimated that identification and removal of colonic adenomas lead to CRC incidence reduction by 25% to 30%.4 One of the most recognized quality indicators of colonoscopy is the adenoma detection rate (ADR).5,6 A greater ADR is associated with longer withdrawal times and increased experience of the endoscopist.7,8 For trainees with limited experience, ADR can remain low despite a long withdrawal time. This underscores the need for an adjunct modality to enhance the ADR of endoscopist trainees.
The main limitation of the ADR is the calculation and reporting hindrance that requires linkage between electronic endoscopic medical reports and pathological report systems for every single polyp removed, which may not be available in all endoscopy units, while the polyp detection rate (PDR) is easier and more practical to retrieve. Several studies have identified a strong association between PDRs and ADRs. Therefore, PDRs have been proposed as ADR surrogate markers, eliminating the need to track final histology.9-12 Recently, advancements in computer-aided polyp detection (CADe) models have shown promising results in the improvement of polyp detection and polyp differentiation.13-18 Therefore, artificial intelligence (AI)-assisted colonoscopy is expected to significantly impact standard endoscopy practices and training.
False positive (FP) alarms are a significant disadvantage of AI-assisted colonoscopies. High false-positive alarm rates can cause stress, visual disturbances, unnecessary checking of non-pathological areas, and prolonged procedure times.19,20 However, lowering the false-positive threshold may also decrease detection sensitivity.21 Thus, we developed “Deep-GI,” an AI model for colonic polyp detection that aimed for a lower FP alarm rate with comparable polyp detection sensitivity (PDS).
The primary objective of our study was to compare the polyp detection performance of "Deep-GI,” a newly developed AI model, to a previously validated CADe (CAD EYE; Fujifilm Corp.) using various FP thresholds, with the secondary goal of determining the best FP threshold for each model, using white light colonoscopies by gastroenterology trainees as a control.
METHODS
Deep-GI model development
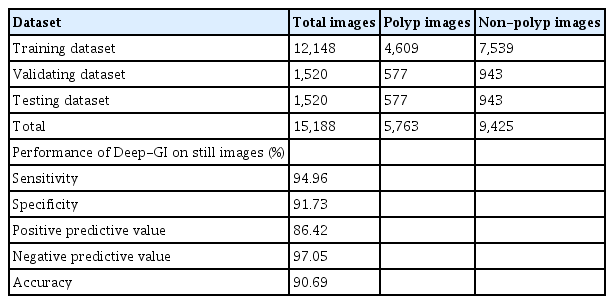
We developed an AI-assisted polyp detection model called “Deep-GI” using colonoscopy images from the Center of Excellence for Innovation and Endoscopy in Gastrointestinal Oncology, King Chulalongkorn Memorial Hospital 2017–2021 database. Both white light endoscopic images and image-enhanced endoscopic images (IEE) using blue light imaging (BLI) and linked color imaging (LCI) were included. All uninformative numerical and nonnumerical data on the captured screen were removed from the raw endoscopic images without any additional modifications or annotations to mimic live endoscopy as much as possible. Two expert endoscopists (KT and SA), each with more than 5 years of experience in colonoscopy and ADR of more than 35% were chosen to review and identify colonic polyps on still images using “LabelMe,” a free open-source labeling software published by Massachusetts Institute of Technology. The labeled images were served as the “ground truth” images. Any discrepancies were resolved by a 3rd expert endoscopist (PM). Following the labeling process, all the images were divided into three datasets. Eighty percent (12,148 images) of the total images were used as the training set, 10% (1,520 images) for internal validation and fine-tuning, and 10% (1,520 images) as the test dataset. The training dataset was subjected to a convolutional neural network, the YOLOv5 deep learning framework, which was specifically designed for real-time detection with an inter-frame space greater than 25 frames per second.22 Supplementary Table 1 provides a detailed description of the still images used in the model.
The Deep-GI model achieved 95% sensitivity, 92% specificity, 86% positive predictive value, 97% negative predictive value, and 91% accuracy, using still images from the test dataset (Table 1).
Performance evaluation
The performance of the Deep-GI model was evaluated using colonoscopy videos. The PDR was compared with that of the trainees, who included five second-year GI fellows with at least 150 colonoscopies performed as the baseline. The performance of the Deep-GI model was also compared to that of a previously validated CADe system (CADe, CAD EYE) using colonoscopy videos in two aspects: (1) PDS and (2) FP alarm rate using various FP thresholds.
Colonoscopy videos were prospectively recorded for participants aged 50 to 75 years who underwent screening colonoscopy at the King Chulalongkorn Memorial Hospital Endoscopy Excellence Center between September 2021 and January 2022. All procedures were performed by gastroenterology trainees with ADRs of ≥35%, under supervision using colonoscopes (ELUXEO 7000 system, EC 760ZP-V/L; Fujifilm Corp.). Patients with a history of CRC, incomplete colonoscopy for inflammatory bowel disease, familial polyposis syndrome, or a history of colonic resection were excluded. Verbal and written informed consent were obtained prior to the procedures. The inspection time was recorded during the withdrawal time, starting from cecal inspection and ending at colonoscope removal from the anus. All colonoscopies were performed under a standard high-definition white light. IEE such as BLI and LCI were only permitted to characterize the polyp. During scope withdrawal, two colonoscopy videos were recorded simultaneously: one with a real-time automatic polyp detection system (CADe)-labeled video and another with an unlabeled raw video. Polypectomy videos were not included in the analysis. The unlabeled video was processed using a Deep-GI model. The same unlabeled videos were also randomly distributed to five independent second-year gastroenterology fellows, who were blinded to the endoscopic and pathological results, to be reviewed to note the number and timing of polyps detected on the screen.
An alarm-tracing program was developed to detect AI-generated frames in the videos. The program was specifically designed to record the number and duration of the appearing bounding boxes regardless of the AI system. This program acted as a “blinded” observer which allowed an objective and reliable measurement of the study outcome. Both the CADe and Deep-GI labeled videos were run through this alarm-tracing program to obtain computerized numbers and durations of the AI-generated blue bounding boxes (Fig. 1).

Example of an adenomatous polyp detected by AI models. (A) A diminutive sessile polyp detected by the computer-aided polyp detection (CADe) model; (B) the same polyp detected by the Deep-GI model; (C) and (D) the alarm-tracing program detecting the bounding boxes of CADe and Deep-GI, respectively.
For expert confirmation or gold standard, three expert gastroenterologists (KT, SA, and PM) independently reviewed both AI-labelled videos. Each expert documented the number and timing of polyps that appeared on the screen as well as their morphology (sessile, pedunculated, or flat) and size (0.5, 0.5–1, or >1 cm). Pathological reports or the reviewers’ consensus (in cases where the polyps were not removed) were used to classify them as adenomatous or hyperplastic.
A true positive (TP) result was defined as a polyp detected for any length of time by the trainees or AI and confirmed by expert reviewers to be a polyp (Fig. 1). A false negative result was defined as a polyp detected by expert reviewers but not by trainees or the AI system. A FP was defined as any area detected by the trainees or the AI system that was not determined to be a polyp by the reviewers. Per-polyp false positivity was used in the study rather than per-frame false positivity for the results to be more clinically relevant. If two frames of the same polyp were deemed to be FP, it was counted as one. Different thresholds for FP alerts were determined based on the length of time for which the system continuously tracked the appearance of FP bounding boxes. The different thresholds of ≥0.5, ≥1, ≥1.5, and ≥2 seconds were adjusted. Finally, the outcomes of all three groups were compared against the gold standard from expert reviewers.
Study-outcomes measurement
Therefore, the primary goal of this study was to compare the polyp detection performance of the Deep-GI model with that of endoscopy trainees and CADe by evaluating the overall PDS and polyp miss rate (PMR). The secondary outcomes were adenoma detection sensitivity (ADS), adenoma miss rate (AMR), and number of FP alarms per colonoscopy using various FP thresholds.
Statistical analysis
According to previous research data on the PDS of recorded trainees, Deep-GI, and published CADe,21 at 80% power and a 2-sided significance level of 0.05, at least 159 videos were required to detect PDS differences. To account for 10% potential exclusions or dropouts, the overall participant enrollment goal was 170.
Analyses were performed using IBM SPSS software ver. 22.0 (IBM Corp.). Categorical variables are expressed as proportions and percentages. Continuous variables are expressed as means and standard deviations. Data between groups were compared using the chi-square test and unpaired t-test, where appropriate. Statistical significance was set at p<0.05. The diagnostic performances of the AI-assisted polyp detection models were expressed in terms of the PDR, PMR, and number of FP alarms per colonoscopy.
Ethical statement
The study protocol was approved by the Institutional Review Board of the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand (IRB number: 56/65). Prior to all procedures, verbal and written informed consent were obtained.
RESULTS
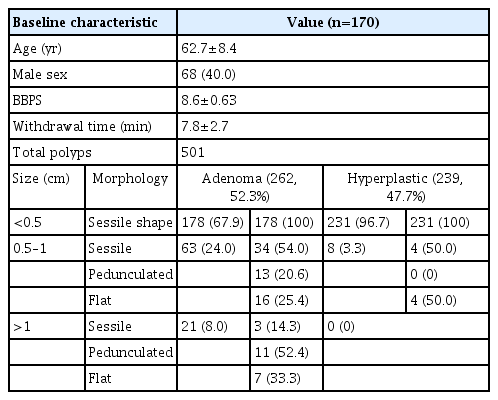
A total of 170 colonoscopies were performed on 68 males (40.0%) and 102 females (60.0%), with a mean age of 62.7±8.4 years. The average withdrawal time was 7.8±2.7 minutes, and the average Boston bowel preparation scale (BPPS) score for bowel preparation quality was 8.6±0.63 points. Of these, 137 patients (80.6%) had at least one polyp. The mean number of polyps detected during colonoscopy was 2.95. A total of 501 polyps were found, of which 262 (52.3%) were adenomas and 239 (47.7%) were hyperplastic polyps.
For adenomatous polyps, majority of the adenomas were <0.5 cm in size (67.9%, n=178). As for the polyp sizes of 0.5 to 0.9 cm, half of them presented sessile morphology (54.0%, n=34). Twenty-one polyps (8%) were >1 cm. The majority of the large polyps were pedunculated (52.4%; n=11). Most of the hyperplastic polyps discovered were <0.5 cm in size (96.7%, n=231). There were also no hyperplastic polyps >1 cm in size or with a pedunculated shape (Table 2).
Polyp detection performance
1) Overall polyp detection
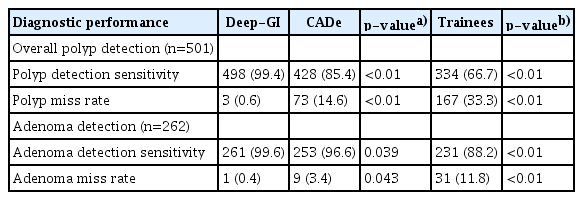
Out of 501 polyps, 498 (99.4%) were detected using Deep-GI. CADe and endoscopist trainees detected 428 (85.4%) and 334 (66.7%) polyps, respectively (p<0.01 for all comparisons). Compared to endoscopist trainees and the validated CADe model, Deep-GI demonstrated a significantly higher PDS with a lower PMR (Table 3).
2) Adenoma detection
Deep-GI detected 261 (99.6%) adenomas, whereas CADe and trainees detected 253 (96.6%) and 231 (88.2%) adenomas out of 262 adenomas, respectively (p<0.01 for all comparisons). Deep-GI demonstrated a significantly higher ADS with a lower AMR than that of trainees and the validated CADe model (Table 3).
3) Missed polyps
Out of 501 polyps detected by the experts, Deep-GI showed the lowest PMR (0.6%, n=3) compared to that of CADe (14.6%, n=73) and trainees (33.3%, n=167), respectively (p<0.01 for all comparisons). Considering only adenomatous polyps, Deep-GI also showed the lowest AMR (0.4%, n=1) compared with that of CADe (3.4%, n=9) and endoscopist trainees (11.8%, n=31), respectively (p<0.01 for all comparisons).
Deep-GI missed one sessile adenoma >1 cm (Fig. 2) and two diminutive hyperplastic polyps, whereas the CADe model missed nine adenomas (including the one missed by Deep-GI) and 64 hyperplastic polyps. The majority of polyps missed by both AI models were <0.5 cm in size. Over 90% of polyps missed by trainees were diminutive (≤5 mm) and most of these polyps (81.4%) were non-neoplastic. The characteristics of all the missed polyps are shown in Supplementary Table 2.
FP alarm rates
Deep-GI displayed 59,350 FP bounding boxes, whereas CADe displayed 106,042 FP bounding boxes in 170 videos. On comparing the two AI systems, Deep-GI showed lower FP alarm rates per colonoscopy (349±169 vs. 624±468, p<0.01). After different FP threshold adjustments, Deep-GI had significantly lower FP alarm rate per colonoscopy than that of CADe at ≥0.5-second and ≥1-second FP thresholds (12.1±10.3 vs. 22.4±23.5, p<0.01 and 4.4±4.8 vs. 6.8±7.6, p<0.01; respectively). However, at a threshold of ≥1.5 and ≥2 seconds, the difference in FP alarm rates became non-significant and the PMR increased to 10% to 25% (Table 4).
DISCUSSION
We discovered that when compared to trainees, the Deep-GI AI model had significantly higher PDS (88% vs. 99%) and ADS (67% vs. 99%), which is consistent with a recent meta-analysis that showed that AI can increase the polyp and adenoma detection by as much as 50%.17 Although our study was designed to blind the subject trainees who reviewed the videos, it had an inevitable limitation in that the trainees had no direct interaction with the AI systems and the effect of incorporating such a system on the trainees’ PDS and ADS could not be proven. However, our findings are strong surrogates, suggesting that AI models have a high potential to improve novice colonoscopy during training.
One of the novelties of this study is the comparison between the newly developed AI model and commercially available systems. With a very high baseline PDS of colonoscopies performed during the study period (>50%), we found that our newly developed AI model, Deep-GI, has a higher sensitivity in polyp detection than that of the commercially available CADe with a sensitivity of 99.4% vs. 85% at the FP threshold of ≥0.5 second and 97.8% vs. 84.2% at the FP threshold of ≥1 second, respectively. When focusing on adenoma detection, the sensitivity of Deep-GI was still higher than that of CADe at the FP thresholds of ≥0.5 second (99.6% vs. 96.2%) and ≥1 second (99.2% vs. 96.2%). Our results demonstrated that Deep-GI performed better than the commercial AI model for overall polyp detection, including adenomas.
Interestingly, only one sessile adenomatous polyp >1 cm was undetected by Deep-GI, CADe, and trainees. We suspect that this large polyp could not be detected for two reasons. First, the polyp was not clearly visible, as it was partially obscured by water and fecal debris, and second, this polyp appeared on the screen for only about 1.5 seconds before polypectomy was performed. Despite these challenges, highly experienced endoscopists were able to detect this polyp during colonoscopy and during offline video assessment.
While an AI system may help improve ADS, high FP alerts may unnecessarily prolong the procedure and increase physical fatigue for the endoscopist. Inevitably, a low FP alert results in lower sensitivity.19,23,24 Our findings highlight the effects of different FP thresholds on the number of FPs reported. Currently, there is no consensus on the optimal FP threshold for AI systems, and various definitions of FP threshold have been used in different CADe studies, ranging from >0.5 to >2 seconds, while some studies have not specified the definition of FP threshold at all.20,25-28 Previous study on another validated polyp detection deep learning AI model (Shanghai Wision AI Co., Ltd.) by Holzwanger et al. proposed ≥2 seconds as the most appropriate and practical threshold for defining FP for colon polyp detection.21 However, in our study, a 2-second threshold resulted in lower PDS and accuracy owing to a higher PMR. In contrast, a ≥1-second threshold provided the lowest PMR while maintaining a low FP alarm. Therefore, we propose an optimum FP threshold of 1 second for the Deep-GI and CADe models, as it provides sufficient time for bubbles or debris to be irrigated away and folds to flatten with insufflation, both of which are standard techniques during high-quality colonoscopy. The different optimal FP observed suggest that the optimal FP threshold for each AI model may be different. However, we believe that a shorter threshold is preferred because the endoscopist does not need to stay in that position for too long.
The strength of this study is that we included a large number of colonoscopy videos with a large number of polyps and adenomas, rendering sufficient power to support the accuracy of our Deep-GI model in terms of PDS. This is the first study to compare and evaluate the diagnostic performance of two different CADe models in terms of PDS and the impact of various FP thresholds. In addition, we evaluated the impact of the time-based definitions of both FP and AI alerts on TP; thus, a sensitivity calculation could be performed accurately.
However, our study has certain limitations. First, the Deep-GI model was not used during real-time colonoscopy, and the benefit of this model in increasing polyp and adenoma detection was only analyzed using offline videos. A randomized controlled trial comparing the two systems in real-time is needed to confirm these findings. Second, Deep-GI was developed, tested, and compared at a single center with no external validation cohort; thus, the superiority of polyp detection results could be due to overfitting or data homogeneity, given the training and testing in the same study population with the same equipment and endoscopists. Third, the CADe system cannot be applied to recorded videos and must be used only during real-time colonoscopy. As a result, the recorded videos may have been influenced by the CADe. Although all annotations were deleted, such as back-to-back colonoscopy, Deep-GI performed better by following and detecting mistakes in prior CADe guidance. In addition, all colonoscopies in this study were performed by endoscopists with high polyp and ADRs under an adequate colonoscopy withdrawal period. In addition, the quality of bowel preparation was excellent in almost all the cases. We did not experience the setting of poor bowel preparation or suboptimal scope withdrawal duration in most cases. In this regard, one advanced adenoma was missed in both AI models owing to debris coverage and a short appearance duration. Therefore, the less-optimal setup may have caused overfitting in our model. Lastly, not all polyp results were based on histopathology, and the Deep-GI capability in differentiating adenomatous vs. non-adenomatous polyps is beyond the scope of our study design, as the main objective of our study was polyp detection, while adenoma detection could be influenced by the proportion of hyperplastic polyps and adenomas in the study population.
In conclusion, on comparing Deep-GI to a validated CADe, Deep-GI demonstrated higher overall PDR and ADR with a significantly lower FP alarm at ≥0.5- and ≥1-second thresholds. The ≥1-second threshold is optimum for Deep-GI model because it provides the lowest PMR and FP alarm rate. To overcome the potential for overfitting, further prospective real-time studies involving community practitioners and trainees are required.
Supplementary Material
Supplementary Table 1. Details of the still images used for Deep-GI development.
Supplementary Table 2. Characteristics of polyps missed by AI systems and endoscopy trainees.
Supplementary materials related to this article can be found online at https://doi.org/10.5946/ce.2023.145.
Notes
Conflicts of Interest
Rungsun Rerknimitr is currently serving as an associate editor in Clinical Endoscopy; however, he was not involved in the peer reviewer selection, evaluation, or decision process for this article. The authors have no potential conflicts of interest.
Funding
This project was funded by the National Research Council of Thailand (NRCT; N42A640330) and the Center of Excellence for Gastrointestinal and Oncology Endoscopy Unit, King Chulalongkorn Memorial Hospital.
Author Contributions
Conceptualization: RR; Data curation: KT, JK, SA, PM, PS, HN, KT, KM, SS, PV; Formal analysis: KT, JK, PM, PS; Funding acquisition: PV, RR; Writing–original draft: KT, JK, PM Writing–review & editing: KT, SA, PM, RR.